Contextual data#
Once you’ve identified areas of interest where multiple datasets intersect, you can pull additional data to provide further context. For example:
landcover
global elevation data
import coincident
import matplotlib.pyplot as plt
import xarray as xr
import numpy as np
import rasterio
/home/docs/checkouts/readthedocs.org/user_builds/coincident/checkouts/104/src/coincident/io/download.py:27: TqdmExperimentalWarning: Using `tqdm.autonotebook.tqdm` in notebook mode. Use `tqdm.tqdm` instead to force console mode (e.g. in jupyter console)
from tqdm.autonotebook import tqdm
%matplotlib inline
# %config InlineBackend.figure_format = 'retina'
Identify a primary dataset#
Start by loading a full resolution polygon of a 3DEP LiDAR workunit which has a known start_datetime and end_datatime:
workunit = "CO_WestCentral_2019"
df_wesm = coincident.search.wesm.read_wesm_csv()
gf_lidar = coincident.search.wesm.load_by_fid(
df_wesm[df_wesm.workunit == workunit].index
)
gf_lidar
| workunit | workunit_id | project | project_id | start_datetime | end_datetime | ql | spec | p_method | dem_gsd_meters | ... | seamless_category | seamless_reason | lpc_link | sourcedem_link | metadata_link | geometry | collection | datetime | dayofyear | duration | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | CO_WestCentral_2019 | 175984 | CO_WestCentral_2019_A19 | 175987 | 2019-08-21 | 2019-09-19 | QL 2 | USGS Lidar Base Specification 1.3 | linear-mode lidar | 1.0 | ... | Meets | Meets 3DEP seamless DEM requirements | https://rockyweb.usgs.gov/vdelivery/Datasets/S... | http://prd-tnm.s3.amazonaws.com/index.html?pre... | http://prd-tnm.s3.amazonaws.com/index.html?pre... | MULTIPOLYGON (((-106.17143 38.42061, -106.3208... | 3DEP | 2019-09-04 12:00:00 | 247 | 29 |
1 rows × 33 columns
search_aoi = gf_lidar.simplify(0.01)
Search for coincident contextual data#
Coincident provides two convenience functions to load datasets resulting from a given search.
gf_wc = coincident.search.search(
dataset="worldcover",
intersects=search_aoi,
# worldcover is just 2020 an 2021, so pick one
datetime=["2020"],
) # Asset of interest = 'map'
# STAC metadata always has a "stac_version" column
gf_wc.iloc[0].stac_version
'1.1.0'
gf_cop30 = coincident.search.search(
dataset="cop30",
intersects=search_aoi,
) # Asset of interest = 'data'
gf_cop30.iloc[0].stac_version
'1.1.0'
STAC search results to datacube#
If the results have STAC-formatted metadata. We can take advantage of the excellent odc.stac tool to load datacubes. Please refer to odc.stac documentation for all the configuration options (e.g. setting resolution our output CRS, etc.)
By default this uses the dask parallel computing library to intelligently load only metadata initially and defer reading values until computations or visualizations are performed.
Copernicus DEM#
ds = coincident.io.xarray.to_dataset(
gf_cop30,
aoi=search_aoi,
# chunks=dict(x=2048, y=2048), # manual chunks
resolution=0.00081, # ~90m
mask=True,
)
# By default, these are dask arrays
ds
<xarray.Dataset> Size: 14MB
Dimensions: (latitude: 1538, longitude: 2288, time: 1)
Coordinates:
* latitude (latitude) float64 12kB 39.39 39.39 39.39 ... 38.15 38.14 38.14
* longitude (longitude) float64 18kB -108.0 -108.0 -108.0 ... -106.1 -106.1
* time (time) datetime64[ns] 8B 2021-04-22
spatial_ref int64 8B 0
Data variables:
data (time, latitude, longitude) float32 14MB dask.array<chunksize=(1, 1538, 2288), meta=np.ndarray># You might want to rename the data variable
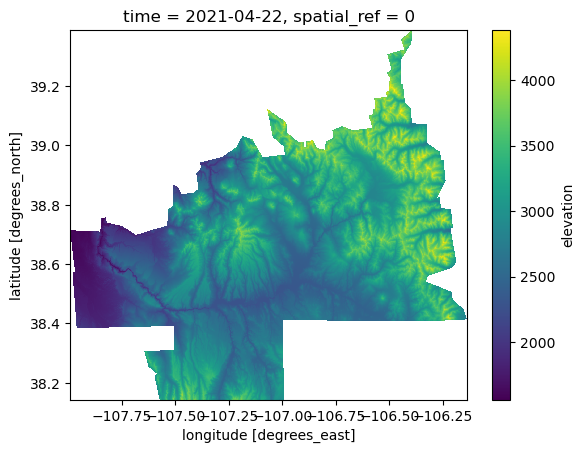
ds = ds.rename(data="elevation")
# The total size of this dataset is only 14MB, so for faster computations, load it into memory
print(ds.nbytes / 1e6)
ds = ds.compute()
14.1064
ds.elevation.isel(time=0).plot.imshow();
ESA Worldcover#
# Same with LandCover
dswc = coincident.io.xarray.to_dataset(
gf_wc,
bands=["map"],
aoi=search_aoi,
mask=True,
# resolution=0.00027, #~30m
resolution=0.00081, # ~90m
)
dswc
<xarray.Dataset> Size: 4MB
Dimensions: (latitude: 1538, longitude: 2288, time: 1)
Coordinates:
* latitude (latitude) float64 12kB 39.39 39.39 39.39 ... 38.15 38.14 38.14
* longitude (longitude) float64 18kB -108.0 -108.0 -108.0 ... -106.1 -106.1
* time (time) datetime64[ns] 8B 2020-01-01
spatial_ref int64 8B 0
Data variables:
map (time, latitude, longitude) uint8 4MB dask.array<chunksize=(1, 1538, 2288), meta=np.ndarray>dswc = dswc.rename(map="landcover")
dswc = dswc.compute()
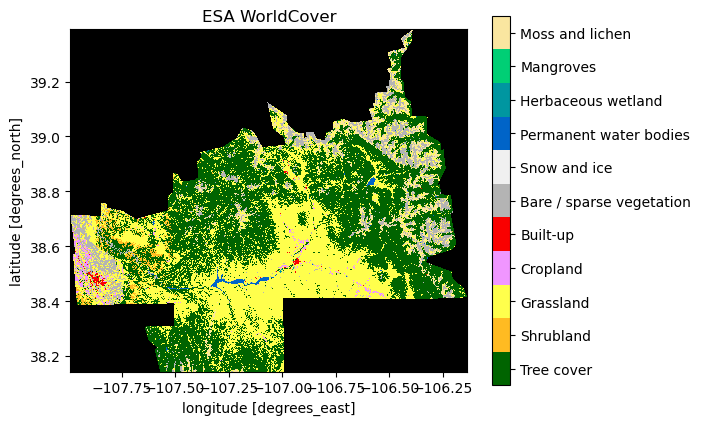
# For landcover there is a convenicence function for a nice categorical colormap
ax = coincident.plot.plot_esa_worldcover(dswc)
ax.set_title("ESA WorldCover");
Load gridded elevation in a consistent CRS#
coincident also has a convenience function for loading gridded elevation datasets in a consistent CRS. In order to facilitate comparison with modern altimetry datasets (ICESat-2, GEDI), we convert elevation data on-the-fly to EPSG:7912. 3D Coordinate Reference Frames are a complex topic, so please see this resource for more detail on how these conversions are done: https://uw-cryo.github.io/3D_CRS_Transformation_Resources/
Note
Currently gridded DEMs are retrieved from OpenTopography hosted in AWS us-west-2.
Warning
The output of load_dem_7912() uses the native resolution and grid of the input dataset, and data is immediately read into memory. So this method is better suited to small AOIs.
# Start with a small AOI:
from shapely.geometry import box
import geopandas as gpd
aoi = gpd.GeoDataFrame(
geometry=[box(-106.812163, 38.40825, -106.396812, 39.049796)], crs="EPSG:4326"
)
da_cop = coincident.io.xarray.load_dem_7912("cop30", aoi=aoi)
da_cop
---------------------------------------------------------------------------
KeyError Traceback (most recent call last)
File ~/checkouts/readthedocs.org/user_builds/coincident/checkouts/104/.pixi/envs/dev/lib/python3.12/site-packages/xarray/backends/file_manager.py:211, in CachingFileManager._acquire_with_cache_info(self, needs_lock)
210 try:
--> 211 file = self._cache[self._key]
212 except KeyError:
File ~/checkouts/readthedocs.org/user_builds/coincident/checkouts/104/.pixi/envs/dev/lib/python3.12/site-packages/xarray/backends/lru_cache.py:56, in LRUCache.__getitem__(self, key)
55 with self._lock:
---> 56 value = self._cache[key]
57 self._cache.move_to_end(key)
KeyError: [<function open at 0x7a22e9441940>, ('https://raw.githubusercontent.com/uw-cryo/3D_CRS_Transformation_Resources/refs/heads/main/globaldems/COP30_hh_7912.vrt',), 'r', (('sharing', False),), 'd4dc1100-518d-4f06-9222-07939777dfd7']
During handling of the above exception, another exception occurred:
CPLE_OpenFailedError Traceback (most recent call last)
File rasterio/_base.pyx:310, in rasterio._base.DatasetBase.__init__()
File rasterio/_base.pyx:221, in rasterio._base.open_dataset()
File rasterio/_err.pyx:359, in rasterio._err.exc_wrap_pointer()
CPLE_OpenFailedError: '/vsicurl/https://raw.githubusercontent.com/uw-cryo/3D_CRS_Transformation_Resources/refs/heads/main/globaldems/COP30_hh_7912.vrt' does not exist in the file system, and is not recognized as a supported dataset name.
During handling of the above exception, another exception occurred:
RasterioIOError Traceback (most recent call last)
Cell In[18], line 1
----> 1 da_cop = coincident.io.xarray.load_dem_7912("cop30", aoi=aoi)
2 da_cop
File ~/checkouts/readthedocs.org/user_builds/coincident/checkouts/104/src/coincident/io/xarray.py:97, in load_dem_7912(dataset, aoi)
94 # Pull all data into memory
95 with ENV:
96 return (
---> 97 xr.open_dataarray(uri, engine="rasterio")
98 .rio.clip_box(**aoi.bounds)
99 .squeeze()
100 .load()
101 )
File ~/checkouts/readthedocs.org/user_builds/coincident/checkouts/104/.pixi/envs/dev/lib/python3.12/site-packages/xarray/backends/api.py:881, in open_dataarray(filename_or_obj, engine, chunks, cache, decode_cf, mask_and_scale, decode_times, decode_timedelta, use_cftime, concat_characters, decode_coords, drop_variables, inline_array, chunked_array_type, from_array_kwargs, backend_kwargs, **kwargs)
710 def open_dataarray(
711 filename_or_obj: str | os.PathLike[Any] | ReadBuffer | AbstractDataStore,
712 *,
(...) 731 **kwargs,
732 ) -> DataArray:
733 """Open an DataArray from a file or file-like object containing a single
734 data variable.
735
(...) 878 open_dataset
879 """
--> 881 dataset = open_dataset(
882 filename_or_obj,
883 decode_cf=decode_cf,
884 mask_and_scale=mask_and_scale,
885 decode_times=decode_times,
886 concat_characters=concat_characters,
887 decode_coords=decode_coords,
888 engine=engine,
889 chunks=chunks,
890 cache=cache,
891 drop_variables=drop_variables,
892 inline_array=inline_array,
893 chunked_array_type=chunked_array_type,
894 from_array_kwargs=from_array_kwargs,
895 backend_kwargs=backend_kwargs,
896 use_cftime=use_cftime,
897 decode_timedelta=decode_timedelta,
898 **kwargs,
899 )
901 if len(dataset.data_vars) != 1:
902 if len(dataset.data_vars) == 0:
File ~/checkouts/readthedocs.org/user_builds/coincident/checkouts/104/.pixi/envs/dev/lib/python3.12/site-packages/xarray/backends/api.py:687, in open_dataset(filename_or_obj, engine, chunks, cache, decode_cf, mask_and_scale, decode_times, decode_timedelta, use_cftime, concat_characters, decode_coords, drop_variables, inline_array, chunked_array_type, from_array_kwargs, backend_kwargs, **kwargs)
675 decoders = _resolve_decoders_kwargs(
676 decode_cf,
677 open_backend_dataset_parameters=backend.open_dataset_parameters,
(...) 683 decode_coords=decode_coords,
684 )
686 overwrite_encoded_chunks = kwargs.pop("overwrite_encoded_chunks", None)
--> 687 backend_ds = backend.open_dataset(
688 filename_or_obj,
689 drop_variables=drop_variables,
690 **decoders,
691 **kwargs,
692 )
693 ds = _dataset_from_backend_dataset(
694 backend_ds,
695 filename_or_obj,
(...) 705 **kwargs,
706 )
707 return ds
File ~/checkouts/readthedocs.org/user_builds/coincident/checkouts/104/.pixi/envs/dev/lib/python3.12/site-packages/rioxarray/xarray_plugin.py:59, in RasterioBackend.open_dataset(self, filename_or_obj, drop_variables, parse_coordinates, lock, masked, mask_and_scale, variable, group, default_name, decode_coords, decode_times, decode_timedelta, band_as_variable, open_kwargs)
57 if open_kwargs is None:
58 open_kwargs = {}
---> 59 rds = _io.open_rasterio(
60 filename_or_obj,
61 parse_coordinates=parse_coordinates,
62 cache=False,
63 lock=lock,
64 masked=masked,
65 mask_and_scale=mask_and_scale,
66 variable=variable,
67 group=group,
68 default_name=default_name,
69 decode_times=decode_times,
70 decode_timedelta=decode_timedelta,
71 band_as_variable=band_as_variable,
72 **open_kwargs,
73 )
74 if isinstance(rds, xarray.DataArray):
75 dataset = rds.to_dataset()
File ~/checkouts/readthedocs.org/user_builds/coincident/checkouts/104/.pixi/envs/dev/lib/python3.12/site-packages/rioxarray/_io.py:1135, in open_rasterio(filename, parse_coordinates, chunks, cache, lock, masked, mask_and_scale, variable, group, default_name, decode_times, decode_timedelta, band_as_variable, **open_kwargs)
1133 else:
1134 manager = URIManager(file_opener, filename, mode="r", kwargs=open_kwargs)
-> 1135 riods = manager.acquire()
1136 captured_warnings = rio_warnings.copy()
1138 # raise the NotGeoreferencedWarning if applicable
File ~/checkouts/readthedocs.org/user_builds/coincident/checkouts/104/.pixi/envs/dev/lib/python3.12/site-packages/xarray/backends/file_manager.py:193, in CachingFileManager.acquire(self, needs_lock)
178 def acquire(self, needs_lock=True):
179 """Acquire a file object from the manager.
180
181 A new file is only opened if it has expired from the
(...) 191 An open file object, as returned by ``opener(*args, **kwargs)``.
192 """
--> 193 file, _ = self._acquire_with_cache_info(needs_lock)
194 return file
File ~/checkouts/readthedocs.org/user_builds/coincident/checkouts/104/.pixi/envs/dev/lib/python3.12/site-packages/xarray/backends/file_manager.py:217, in CachingFileManager._acquire_with_cache_info(self, needs_lock)
215 kwargs = kwargs.copy()
216 kwargs["mode"] = self._mode
--> 217 file = self._opener(*self._args, **kwargs)
218 if self._mode == "w":
219 # ensure file doesn't get overridden when opened again
220 self._mode = "a"
File ~/checkouts/readthedocs.org/user_builds/coincident/checkouts/104/.pixi/envs/dev/lib/python3.12/site-packages/rasterio/env.py:463, in ensure_env_with_credentials.<locals>.wrapper(*args, **kwds)
460 session = DummySession()
462 with env_ctor(session=session):
--> 463 return f(*args, **kwds)
File ~/checkouts/readthedocs.org/user_builds/coincident/checkouts/104/.pixi/envs/dev/lib/python3.12/site-packages/rasterio/__init__.py:356, in open(fp, mode, driver, width, height, count, crs, transform, dtype, nodata, sharing, opener, **kwargs)
353 path = _parse_path(raw_dataset_path)
355 if mode == "r":
--> 356 dataset = DatasetReader(path, driver=driver, sharing=sharing, **kwargs)
357 elif mode == "r+":
358 dataset = get_writer_for_path(path, driver=driver)(
359 path, mode, driver=driver, sharing=sharing, **kwargs
360 )
File rasterio/_base.pyx:312, in rasterio._base.DatasetBase.__init__()
RasterioIOError: '/vsicurl/https://raw.githubusercontent.com/uw-cryo/3D_CRS_Transformation_Resources/refs/heads/main/globaldems/COP30_hh_7912.vrt' does not exist in the file system, and is not recognized as a supported dataset name.
# NASA DEM and COP30 DEM are both 30m but will not necessarily have same coordinates!
da_nasa = coincident.io.xarray.load_dem_7912("nasadem", aoi=aoi)
da_nasa
# We check that coordinates are sufficiently close before force-alignment.
# If sufficiently different, it would be better to use rioxarray.reproject_match!
xr.testing.assert_allclose(da_cop.x, da_nasa.x) # rtol=1e-05, atol=1e-08 defaults
xr.testing.assert_allclose(da_cop.y, da_nasa.y)
da_nasa = da_nasa.assign_coords(x=da_cop.x, y=da_cop.y)
diff = da_cop - da_nasa
median = diff.median()
mean = diff.mean()
std = diff.std()
fig = plt.figure(layout="constrained", figsize=(4, 8)) # figsize=(8.5, 11)
ax = fig.add_gridspec(top=0.75).subplots()
ax_histx = ax.inset_axes([0, -0.35, 1, 0.25])
axes = [ax, ax_histx]
diff.plot.imshow(
ax=axes[0], robust=True, add_colorbar=False
) # , cbar_kwargs={'label': ""})
n, bins, ax = diff.plot.hist(ax=axes[1], bins=100, range=(-20, 20), color="gray")
approx_mode = bins[np.argmax(n)]
axes[0].set_title("COP30 - NASADEM")
axes[0].set_aspect(aspect=1 / np.cos(np.deg2rad(38.7)))
axes[1].axvline(0, color="k")
axes[1].axvline(median, label=f"median={median:.2f}", color="cyan", lw=1)
axes[1].axvline(mean, label=f"mode={mean:.2f}", color="magenta", lw=1)
axes[1].axvline(approx_mode, label=f"mode={approx_mode:.2f}", color="yellow", lw=1)
axes[1].set_xlabel("Elevation difference (m)")
axes[1].legend()
axes[1].set_title("");
Note
We see that COP30 elevation values are approximately 1m greater than NASADEM values for this area. Such a difference is within the stated accuracies of each gridded dataset, and also COP30 is derived from X-band TanDEM-X observations with nominal time of 2021-04-22 whereas NASADEM is primarily based on C-band SRTM collected 2000-02-20.
# Load 3DEP 10m data
da_3dep = coincident.io.xarray.load_dem_7912("3dep", aoi=aoi)
# To compare to 3dep, we refine cop30 from 30m to 10m using bilinear resampling
da_cop_r = da_cop.rio.reproject_match(
da_3dep, resampling=rasterio.enums.Resampling.bilinear
)
da_cop_r
diff = da_3dep - da_cop_r
median = diff.median()
mean = diff.mean()
std = diff.std()
fig = plt.figure(layout="constrained", figsize=(4, 8)) # figsize=(8.5, 11)
ax = fig.add_gridspec(top=0.75).subplots()
ax_histx = ax.inset_axes([0, -0.35, 1, 0.25])
axes = [ax, ax_histx]
diff.plot.imshow(
ax=axes[0], robust=True, add_colorbar=False
) # , cbar_kwargs={'label': ""})
n, bins, ax = diff.plot.hist(ax=axes[1], bins=100, range=(-20, 20), color="gray")
approx_mode = bins[np.argmax(n)]
axes[0].set_title("3DEP - COP30")
axes[0].set_aspect(aspect=1 / np.cos(np.deg2rad(38.7)))
axes[1].axvline(0, color="k")
axes[1].axvline(median, label=f"median={median:.2f}", color="cyan", lw=1)
axes[1].axvline(mean, label=f"mode={mean:.2f}", color="magenta", lw=1)
axes[1].axvline(approx_mode, label=f"mode={approx_mode:.2f}", color="yellow", lw=1)
axes[1].set_xlabel("Elevation difference (m)")
axes[1].legend()
axes[1].set_title("");
Note
There is a clear spatial and terrain dependence for the residuals of 10m 3DEP LiDAR compared to COP30 elevation values for this area!