Reading and Downloading DEM and LPC Products#
The coincident.io.xarray and coincident.io.download modules support the option to load DEMs into memory via odc-stac or download DEMs into local directories.
There is also support for Lidar Point Cloud (LPC) spatial filtering for aerial lidar catalogs, where the user can return a GeoDataFrame with the respective .laz tile filename, download url, and geometry (epsg 4326) for each tile intersecting an input aoi. These laz files can then be downloaded locally with coincident.io.download.download_files()
There is specific support for USGS 3DEP EPT readers where the user can return a PDAL pipeline configured with the EPT URL, the AOI’s bounds, and polygon WKT, all in the EPT’s spatial reference system.
Note
Coincident does not support the processing of lidar point cloud products. Please see the lidar_tools repository for information on processing the returned GeoDataFrame with lidar point cloud products.
import coincident
import geopandas as gpd
from shapely.geometry import box
/home/docs/checkouts/readthedocs.org/user_builds/coincident/checkouts/104/src/coincident/io/download.py:27: TqdmExperimentalWarning: Using `tqdm.autonotebook.tqdm` in notebook mode. Use `tqdm.tqdm` instead to force console mode (e.g. in jupyter console)
from tqdm.autonotebook import tqdm
3DEP and NEON overlapping Flights#
Note
For all of these functions, you will need identification metadata from the coincident.search.search functions for each respective catalog
Search#
workunit = "CO_CentralEasternPlains_1_2020"
df_wesm = coincident.search.wesm.read_wesm_csv()
gf_usgs = coincident.search.wesm.load_by_fid(
df_wesm[df_wesm.workunit == workunit].index
)
gf_usgs
| workunit | workunit_id | project | project_id | start_datetime | end_datetime | ql | spec | p_method | dem_gsd_meters | ... | seamless_category | seamless_reason | lpc_link | sourcedem_link | metadata_link | geometry | collection | datetime | dayofyear | duration | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | CO_CentralEasternPlains_1_2020 | 192973 | CO_CentralEasternPlains_2020_D20 | 192976 | 2020-05-09 | 2020-06-10 | QL 2 | USGS Lidar Base Specification 2.1 | linear-mode lidar | 1.0 | ... | Meets | Meets 3DEP seamless DEM requirements | https://rockyweb.usgs.gov/vdelivery/Datasets/S... | http://prd-tnm.s3.amazonaws.com/index.html?pre... | http://prd-tnm.s3.amazonaws.com/index.html?pre... | MULTIPOLYGON (((-103.7138 39.8837, -103.7138 3... | 3DEP | 2020-05-25 | 146 | 32 |
1 rows × 33 columns
# We will examine the 'sourcedem' 1m
# NOTE: it's important to note the *vertical* CRS of the data
gf_usgs.iloc[0]
workunit CO_CentralEasternPlains_1_2020
workunit_id 192973
project CO_CentralEasternPlains_2020_D20
project_id 192976
start_datetime 2020-05-09 00:00:00
end_datetime 2020-06-10 00:00:00
ql QL 2
spec USGS Lidar Base Specification 2.1
p_method linear-mode lidar
dem_gsd_meters 1.0
horiz_crs 6342
vert_crs 5703
geoid GEOID18
lpc_pub_date 2020-12-11 00:00:00
lpc_update NaT
lpc_category Meets
lpc_reason Meets 3DEP LPC requirements
sourcedem_pub_date 2020-12-10 00:00:00
sourcedem_update NaT
sourcedem_category Meets
sourcedem_reason Meets 3DEP source DEM requirements
onemeter_category Meets
onemeter_reason Meets 3DEP 1-m DEM requirements
seamless_category Meets
seamless_reason Meets 3DEP seamless DEM requirements
lpc_link https://rockyweb.usgs.gov/vdelivery/Datasets/S...
sourcedem_link http://prd-tnm.s3.amazonaws.com/index.html?pre...
metadata_link http://prd-tnm.s3.amazonaws.com/index.html?pre...
geometry MULTIPOLYGON (((-103.7138 39.8837, -103.7138 3...
collection 3DEP
datetime 2020-05-25 00:00:00
dayofyear 146
duration 32
Name: 0, dtype: object
Now, we’ll explore an overlapping NEON flight
gf_neon = coincident.search.search(
dataset="neon", intersects=gf_usgs, datetime=["2020"]
)
/home/docs/checkouts/readthedocs.org/user_builds/coincident/checkouts/104/.pixi/envs/dev/lib/python3.12/site-packages/geopandas/array.py:403: UserWarning: Geometry is in a geographic CRS. Results from 'sjoin_nearest' are likely incorrect. Use 'GeoSeries.to_crs()' to re-project geometries to a projected CRS before this operation.
warnings.warn(
gf_neon.head()
| id | title | start_datetime | end_datetime | product_url | geometry | |
|---|---|---|---|---|---|---|
| 0 | ARIK | Arikaree River NEON | 2020-06-01 | 2020-06-30 | https://data.neonscience.org/api/v0/data/DP3.3... | POLYGON ((-102.60902 39.69825, -102.60871 39.7... |
m = gf_usgs.explore(color="black")
gf_neon.explore(m=m, column="id", popups=True)
Subset data#
We will evaluate small subset of this overlap for deomstrative purposes.
Let’s subset based on some contextual LULC data
gf_wc = coincident.search.search(
dataset="worldcover",
intersects=gf_neon,
datetime=["2020"],
)
dswc = coincident.io.xarray.to_dataset(
gf_wc,
bands=["map"],
aoi=gf_neon,
mask=True,
resolution=0.00027, # ~30m
)
dswc = dswc.rename(map="landcover")
dswc = dswc.compute()
# arbitrary bbox that will be our subset area (all cropland)
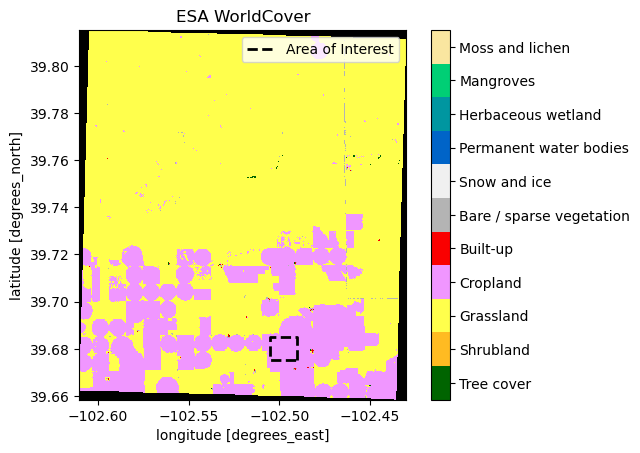
bbox_geometry = box(-102.505, 39.675, -102.49, 39.685)
aoi = gpd.GeoDataFrame(geometry=[bbox_geometry], crs="EPSG:4326")
ax = coincident.plot.plot_esa_worldcover(dswc)
aoi.plot(ax=ax, facecolor="none", edgecolor="black", linestyle="--", linewidth=2)
from matplotlib.lines import Line2D
custom_line = Line2D([0], [0], color="black", linestyle="--", lw=2)
ax.legend([custom_line], ["Area of Interest"], loc="upper right", fontsize=10)
ax.set_title("ESA WorldCover");
Actually read in the DEMs
datetime_str = gf_neon.end_datetime.item()
site_id = gf_neon.id.item()
datetime_str, site_id
('2020-06-30', 'ARIK')
%%time
da_neon_dem = coincident.io.xarray.load_neon_dem(
aoi, datetime_str=datetime_str, site_id=site_id, product="dsm"
)
/home/docs/checkouts/readthedocs.org/user_builds/coincident/checkouts/104/.pixi/envs/dev/lib/python3.12/site-packages/rioxarray/_io.py:1146: RuntimeWarning: TIFFReadDirectoryCheckOrder:Invalid TIFF directory; tags are not sorted in ascending order
if riods.subdatasets:
/home/docs/checkouts/readthedocs.org/user_builds/coincident/checkouts/104/.pixi/envs/dev/lib/python3.12/site-packages/rioxarray/_io.py:1146: RuntimeWarning: TIFFReadDirectoryCheckOrder:Invalid TIFF directory; tags are not sorted in ascending order
if riods.subdatasets:
/home/docs/checkouts/readthedocs.org/user_builds/coincident/checkouts/104/.pixi/envs/dev/lib/python3.12/site-packages/rioxarray/_io.py:1146: RuntimeWarning: TIFFReadDirectoryCheckOrder:Invalid TIFF directory; tags are not sorted in ascending order
if riods.subdatasets:
/home/docs/checkouts/readthedocs.org/user_builds/coincident/checkouts/104/.pixi/envs/dev/lib/python3.12/site-packages/rioxarray/_io.py:1146: RuntimeWarning: TIFFReadDirectoryCheckOrder:Invalid TIFF directory; tags are not sorted in ascending order
if riods.subdatasets:
/home/docs/checkouts/readthedocs.org/user_builds/coincident/checkouts/104/.pixi/envs/dev/lib/python3.12/site-packages/rioxarray/_io.py:1146: RuntimeWarning: TIFFReadDirectoryCheckOrder:Invalid TIFF directory; tags are not sorted in ascending order
if riods.subdatasets:
CPU times: user 181 ms, sys: 33.2 ms, total: 214 ms
Wall time: 1.35 s
/home/docs/checkouts/readthedocs.org/user_builds/coincident/checkouts/104/.pixi/envs/dev/lib/python3.12/site-packages/rioxarray/_io.py:1146: RuntimeWarning: TIFFReadDirectoryCheckOrder:Invalid TIFF directory; tags are not sorted in ascending order
if riods.subdatasets:
da_neon_dem
<xarray.DataArray 'elevation' (y: 1146, x: 1318)> Size: 6MB
dask.array<getitem, shape=(1146, 1318), dtype=float32, chunksize=(808, 1000), chunktype=numpy.ndarray>
Coordinates:
* x (x) float64 11kB 7.14e+05 7.14e+05 ... 7.153e+05 7.153e+05
* y (y) float64 9kB 4.395e+06 4.395e+06 ... 4.396e+06 4.396e+06
spatial_ref int64 8B 0usgs_project = gf_usgs["project"].item()
usgs_project
'CO_CentralEasternPlains_2020_D20'
%%time
da_usgs_dem = coincident.io.xarray.load_usgs_dem(aoi, usgs_project)
CPU times: user 576 ms, sys: 618 ms, total: 1.19 s
Wall time: 2.81 s
da_usgs_dem
<xarray.DataArray 'elevation' (y: 1146, x: 1317)> Size: 6MB
dask.array<getitem, shape=(1146, 1317), dtype=float32, chunksize=(1146, 1317), chunktype=numpy.ndarray>
Coordinates:
* x (x) float64 11kB 7.14e+05 7.14e+05 ... 7.153e+05 7.153e+05
* y (y) float64 9kB 4.396e+06 4.396e+06 ... 4.395e+06 4.395e+06
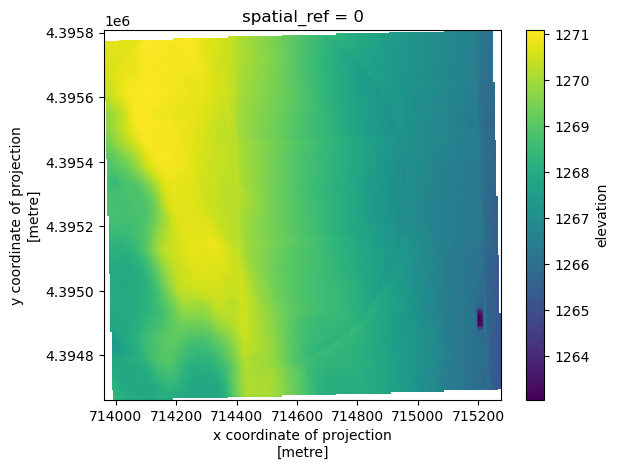
spatial_ref int64 8B 0da_usgs_dem.coarsen(x=5, y=5, boundary="trim").mean().plot.imshow();
Download#
Note
coincident.io.download.download_neon_dem needs the NEON site’s start_datetime OR end_datetime to work
gf_neon
| id | title | start_datetime | end_datetime | product_url | geometry | |
|---|---|---|---|---|---|---|
| 0 | ARIK | Arikaree River NEON | 2020-06-01 | 2020-06-30 | https://data.neonscience.org/api/v0/data/DP3.3... | POLYGON ((-102.60902 39.69825, -102.60871 39.7... |
local_output_dir = "/tmp"
coincident.io.download.download_neon_dem(
aoi=aoi,
datetime_str=gf_neon.start_datetime.item(),
site_id=gf_neon.id.item(),
product="dsm",
output_dir=local_output_dir,
)
# USGS_1M_13_x71y440_CO_CentralEasternPlains_2020_D20.tif: 236MB
coincident.io.download.download_usgs_dem(
aoi=aoi,
project=usgs_project,
output_dir=local_output_dir,
save_parquet=True, # save a STAC-like geoparquet of the tiles you download
)
Finally, you can grab the LPC tile metadata. For USGS 3DEP data, you can also return a PDAL pipeline based on the available EPT data. This PDAL pipeline will be returned as a JSON file where the user can add their own custom parameters (additional filters, writers, etc.) to this pipeline dictionary before executing it with PDAL.
Note
coincident.io.download.fetch_lpc_tiles needs the NEON site’s end_datetime OR start_datetime to work if you are in fact accessing NEON data
%%time
gf_neon_lpc_tiles = coincident.io.download.fetch_lpc_tiles(
aoi=aoi,
dataset_id=gf_neon.id.item(),
provider="NEON",
datetime_str=gf_neon.end_datetime.item(),
)
CPU times: user 53.5 ms, sys: 5.14 ms, total: 58.7 ms
Wall time: 329 ms
gf_neon_lpc_tiles.head()
| name | url | geometry | |
|---|---|---|---|
| 0 | NEON_D10_ARIK_DP1_715000_4395000_classified_po... | https://storage.googleapis.com/neon-aop-produc... | POLYGON ((-102.48149 39.67754, -102.48116 39.6... |
| 1 | NEON_D10_ARIK_DP1_714000_4394000_classified_po... | https://storage.googleapis.com/neon-aop-produc... | POLYGON ((-102.49347 39.66879, -102.49314 39.6... |
| 2 | NEON_D10_ARIK_DP1_713000_4395000_classified_po... | https://storage.googleapis.com/neon-aop-produc... | POLYGON ((-102.50479 39.67804, -102.50447 39.6... |
| 3 | NEON_D10_ARIK_DP1_713000_4394000_classified_po... | https://storage.googleapis.com/neon-aop-produc... | POLYGON ((-102.50511 39.66904, -102.50479 39.6... |
| 4 | NEON_D10_ARIK_DP1_714000_4395000_classified_po... | https://storage.googleapis.com/neon-aop-produc... | POLYGON ((-102.49314 39.67779, -102.49281 39.6... |
%%time
gf_usgs_lpc_tiles = coincident.io.download.fetch_lpc_tiles(
aoi=aoi, dataset_id=usgs_project, provider="USGS"
)
CPU times: user 10.2 ms, sys: 0 ns, total: 10.2 ms
Wall time: 1.37 s
gf_usgs_lpc_tiles.head()
| name | url | geometry | |
|---|---|---|---|
| 0 | 5fded821d34e30b9123e230c | https://rockyweb.usgs.gov/vdelivery/Datasets/S... | POLYGON ((-102.50479 39.66904, -102.50479 39.6... |
| 1 | 5fded821d34e30b9123e230e | https://rockyweb.usgs.gov/vdelivery/Datasets/S... | POLYGON ((-102.50447 39.67804, -102.50447 39.6... |
| 2 | 5fded82fd34e30b9123e235a | https://rockyweb.usgs.gov/vdelivery/Datasets/S... | POLYGON ((-102.49314 39.66879, -102.49314 39.6... |
| 3 | 5fded830d34e30b9123e235c | https://rockyweb.usgs.gov/vdelivery/Datasets/S... | POLYGON ((-102.49281 39.67779, -102.49281 39.6... |
| 4 | 5fded837d34e30b9123e23a8 | https://rockyweb.usgs.gov/vdelivery/Datasets/S... | POLYGON ((-102.48149 39.66854, -102.48149 39.6... |
m = gf_usgs_lpc_tiles.explore(color="black")
gf_neon_lpc_tiles.explore(m=m)
Now, we can download the laz files
coincident.io.download.download_files(
gf_neon_lpc_tiles["url"], output_dir=local_output_dir
)
pdal_pipeline = coincident.io.download.build_usgs_ept_pipeline(
aoi, workunit=gf_usgs.workunit.item(), output_dir=local_output_dir
)
pdal_pipeline
{'pipeline': [{'type': 'readers.ept',
'filename': 'https://s3-us-west-2.amazonaws.com/usgs-lidar-public/CO_CentralEasternPlains_1_2020/ept.json',
'bounds': '(([-11410804.4037645068, -11409134.6114026085], [4818825.9525320893, 4820272.3693662276]))'},
{'type': 'filters.crop',
'polygon': 'POLYGON ((-11409134.611402608 4818825.952532089, -11409134.611402608 4820272.369366228, -11410804.403764507 4820272.369366228, -11410804.403764507 4818825.952532089, -11409134.611402608 4818825.952532089))'},
{'type': 'writers.las',
'filename': 'CO_CentralEasternPlains_1_2020_EPT_subset_pipeline.laz',
'compression': 'laszip'}]}
NCALM#
Search#
aoi = gpd.read_file(
"https://raw.githubusercontent.com/unitedstates/districts/refs/heads/gh-pages/states/WA/shape.geojson"
)
aoi.plot();
gf_ncalm = coincident.search.search(
dataset="ncalm", intersects=aoi, datetime=["2018-09-19"]
)
gf_ncalm
| id | name | title | start_datetime | end_datetime | geometry | |
|---|---|---|---|---|---|---|
| 0 | OTLAS.072019.6339.1 | WA18_Wall | High-Resolution Mapping of Goat Rock Volcano, WA | 2018-09-19 | 2018-09-20 | POLYGON ((-121.46701 46.48376, -121.45914 46.4... |
Now, let’s subset to a small AOI for convenience sake
buffer_size = 0.01
centroid = gf_ncalm.centroid
mini_aoi = gpd.GeoDataFrame(
geometry=[
box(
centroid.x - buffer_size,
centroid.y - buffer_size,
centroid.x + buffer_size,
centroid.y + buffer_size,
)
],
crs="EPSG:4326",
)
/tmp/ipykernel_1139/991173689.py:2: UserWarning: Geometry is in a geographic CRS. Results from 'centroid' are likely incorrect. Use 'GeoSeries.to_crs()' to re-project geometries to a projected CRS before this operation.
centroid = gf_ncalm.centroid
/home/docs/checkouts/readthedocs.org/user_builds/coincident/checkouts/104/.pixi/envs/dev/lib/python3.12/site-packages/shapely/geometry/polygon.py:91: FutureWarning: Calling float on a single element Series is deprecated and will raise a TypeError in the future. Use float(ser.iloc[0]) instead
return [float(c) for c in o]
m = gf_ncalm.explore()
mini_aoi.explore(m=m, color="red")
Note
The NCALM DEMs are not tiled, so reading in the data and downloading takes a longer time
Subset data#
%%time
da_ncalm_dtm = coincident.io.xarray.load_ncalm_dem(
aoi=mini_aoi, product="dtm", dataset_id=gf_ncalm["name"].item()
)
CPU times: user 6.36 s, sys: 3.58 s, total: 9.94 s
Wall time: 23.5 s
da_ncalm_dtm
<xarray.DataArray 'elevation' (y: 2254, x: 1579)> Size: 14MB
array([[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan],
...,
[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan]],
shape=(2254, 1579), dtype=float32)
Coordinates:
* x (x) float64 13kB 6.226e+05 6.226e+05 ... 6.242e+05 6.242e+05
* y (y) float64 18kB 5.152e+06 5.152e+06 ... 5.15e+06 5.15e+06
spatial_ref int64 8B 0
Attributes:
AREA_OR_POINT: Area
scale_factor: 1.0
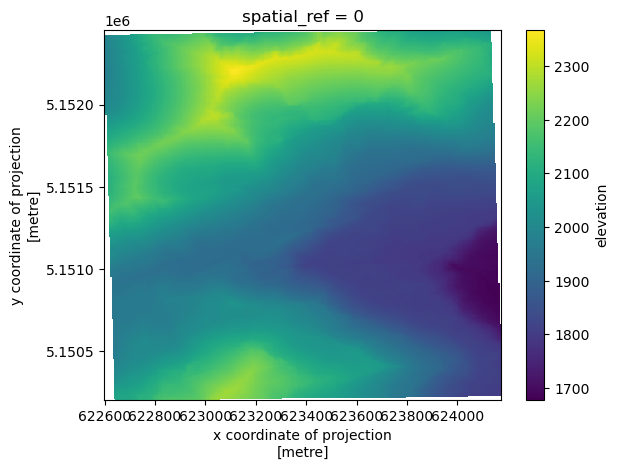
add_offset: 0.0da_ncalm_dtm.coarsen(x=5, y=5, boundary="trim").mean().plot.imshow();
Download#
Note
You will not see a download progress bar for downloading NCALM DEMs as opposed to USGS and NEON provider DEMs
%%time
coincident.io.download.download_ncalm_dem(
aoi=mini_aoi,
dataset_id=gf_ncalm["name"].item(),
product="dtm",
output_dir=local_output_dir,
)
CPU times: user 6.18 s, sys: 1.64 s, total: 7.81 s
Wall time: 20.6 s
%%time
gf_ncalm_lpc_tiles = coincident.io.download.fetch_lpc_tiles(
aoi=mini_aoi, dataset_id=gf_ncalm.name.item(), provider="NCALM"
)
CPU times: user 64.3 ms, sys: 5.45 ms, total: 69.7 ms
Wall time: 449 ms
gf_ncalm_lpc_tiles.head()
| name | url | geometry | |
|---|---|---|---|
| 0 | WA18_Wall/622000_5150000.laz | https://opentopography.s3.sdsc.edu/pc-bulk/WA1... | POLYGON ((-121.39723 46.49234, -121.39697 46.5... |
| 1 | WA18_Wall/622000_5151000.laz | https://opentopography.s3.sdsc.edu/pc-bulk/WA1... | POLYGON ((-121.39697 46.50133, -121.3967 46.51... |
| 2 | WA18_Wall/622000_5152000.laz | https://opentopography.s3.sdsc.edu/pc-bulk/WA1... | POLYGON ((-121.3967 46.51033, -121.39644 46.51... |
| 3 | WA18_Wall/623000_5150000.laz | https://opentopography.s3.sdsc.edu/pc-bulk/WA1... | POLYGON ((-121.3842 46.49216, -121.38394 46.50... |
| 4 | WA18_Wall/623000_5151000.laz | https://opentopography.s3.sdsc.edu/pc-bulk/WA1... | POLYGON ((-121.38394 46.50115, -121.38367 46.5... |
m = gf_ncalm.explore()
gf_ncalm_lpc_tiles.explore(m=m, color="red")
mini_aoi.explore(m=m, color="black")
NOAA#
Search#
aoi = gpd.read_file(
"https://raw.githubusercontent.com/unitedstates/districts/refs/heads/gh-pages/states/FL/shape.geojson"
)
gf_noaa = coincident.search.search(
dataset="noaa", intersects=aoi, datetime=["2022-10-27"]
)
buffer_size = 0.02
centroid = gf_noaa.centroid
mini_aoi = gpd.GeoDataFrame(
geometry=[
box(
centroid.x - buffer_size,
centroid.y - buffer_size,
centroid.x + buffer_size,
centroid.y + buffer_size,
)
],
crs="EPSG:4326",
)
/tmp/ipykernel_1139/2106933600.py:2: UserWarning: Geometry is in a geographic CRS. Results from 'centroid' are likely incorrect. Use 'GeoSeries.to_crs()' to re-project geometries to a projected CRS before this operation.
centroid = gf_noaa.centroid
/home/docs/checkouts/readthedocs.org/user_builds/coincident/checkouts/104/.pixi/envs/dev/lib/python3.12/site-packages/shapely/geometry/polygon.py:91: FutureWarning: Calling float on a single element Series is deprecated and will raise a TypeError in the future. Use float(ser.iloc[0]) instead
return [float(c) for c in o]
m = gf_noaa.explore()
mini_aoi.explore(m=m, color="red")
the name and id being identical is expected
gf_noaa
| id | name | title | start_datetime | end_datetime | geometry | |
|---|---|---|---|---|---|---|
| 0 | 10149 | 10149 | 2022 NOAA NGS Topobathy Lidar: Big Bend WMA, FL | 2022-10-27 | 2022-11-29 | POLYGON ((-83.4654 29.36713, -83.35291 29.4196... |
Note
Our coincident.search.search(dataset=”noaa”) returns the dataset ids “Lidar Datasets at NOAA Digital Coast” whereas coincident.io.xarray.load_noaa_dem() requires the ids from the “Imagery and Elevation Raster Datasets at NOAA Digital Coast” dataset. The corresponding elevation raster dataset id is the same as the lidar dataset id + 1. e.g. “Great Bay NERR UAS Lidar” has id 10175 for lidar data and id 10176 for dem data
noaa_dem_id = int(gf_noaa.id.item()) + 1
print(f"NOAA LiDAR id: {gf_noaa.id.item()} NOAA DEM id: {noaa_dem_id}")
NOAA LiDAR id: 10149 NOAA DEM id: 10150
Warning
The larger the NOAA flight, the longer the below function takes regardless of your input AOI
Subset#
%%time
da_noaa_dem = coincident.io.xarray.load_noaa_dem(mini_aoi, noaa_dem_id)
CPU times: user 1.67 s, sys: 282 ms, total: 1.96 s
Wall time: 3.69 s
da_noaa_dem
<xarray.DataArray 'elevation' (y: 3909, x: 3964)> Size: 62MB
dask.array<getitem, shape=(3909, 3964), dtype=float32, chunksize=(3909, 2625), chunktype=numpy.ndarray>
Coordinates:
* x (x) float64 32kB 2.237e+05 2.237e+05 ... 2.276e+05 2.276e+05
* y (y) float64 31kB 3.304e+06 3.304e+06 ... 3.3e+06 3.3e+06
spatial_ref int64 8B 0da_noaa_dem.coarsen(x=50, y=50, boundary="trim").mean().plot.imshow();
buffer_size = 0.008
centroid = gf_noaa.centroid
mini_aoi = gpd.GeoDataFrame(
geometry=[
box(
centroid.x - buffer_size,
centroid.y - buffer_size,
centroid.x + buffer_size,
centroid.y + buffer_size,
)
],
crs="EPSG:4326",
)
m = gf_noaa.explore()
mini_aoi.explore(m=m, color="red")
/tmp/ipykernel_1139/1549685662.py:2: UserWarning: Geometry is in a geographic CRS. Results from 'centroid' are likely incorrect. Use 'GeoSeries.to_crs()' to re-project geometries to a projected CRS before this operation.
centroid = gf_noaa.centroid
/home/docs/checkouts/readthedocs.org/user_builds/coincident/checkouts/104/.pixi/envs/dev/lib/python3.12/site-packages/shapely/geometry/polygon.py:91: FutureWarning: Calling float on a single element Series is deprecated and will raise a TypeError in the future. Use float(ser.iloc[0]) instead
return [float(c) for c in o]
Download#
Note
You will not see a download progress bar for downloading NOAA DEMs as opposed to USGS and NEON provider DEMs
%%time
coincident.io.download.download_noaa_dem(
aoi=mini_aoi, dataset_id=noaa_dem_id, output_dir=local_output_dir
)
CPU times: user 2.26 s, sys: 491 ms, total: 2.76 s
Wall time: 3.71 s
list(mini_aoi.bounds.values)
[array([-83.84683768, 29.80678818, -83.83083768, 29.82278818])]
gf_noaa.id.item()
'10149'
%%time
gf_noaa_lpc_tiles = coincident.io.download.fetch_lpc_tiles(
aoi=mini_aoi, dataset_id=gf_noaa.id.item(), provider="NOAA"
)
CPU times: user 1.86 s, sys: 26.8 ms, total: 1.88 s
Wall time: 3.73 s
gf_noaa_lpc_tiles
| name | url | geometry | |
|---|---|---|---|
| 0 | 20221114_225500e_3302500n.copc.laz | https://noaa-nos-coastal-lidar-pds.s3.amazonaw... | POLYGON ((-83.8353 29.82259, -83.83543 29.8270... |
| 1 | 20221114_226000e_3302000n.copc.laz | https://noaa-nos-coastal-lidar-pds.s3.amazonaw... | POLYGON ((-83.83 29.81819, -83.83013 29.8227, ... |
| 2 | 20221114_226000e_3302500n.copc.laz | https://noaa-nos-coastal-lidar-pds.s3.amazonaw... | POLYGON ((-83.83013 29.8227, -83.83026 29.8272... |
m = gf_noaa.explore()
gf_noaa_lpc_tiles.explore(m=m, color="red")
G-LiHT#
Search#
aoi = gpd.read_file(
"https://raw.githubusercontent.com/unitedstates/districts/refs/heads/gh-pages/states/MD/shape.geojson"
)
aoi = aoi.simplify(0.01)
aoi.explore()
gf_gliht = coincident.search.search(
dataset="gliht",
intersects=aoi,
datetime=["2017-07-31"],
)
gf_gliht = gf_gliht.iloc[[0]]
gf_gliht
| assets | bbox | collection | geometry | id | links | stac_extensions | stac_version | type | datetime | end_datetime | start_datetime | storage:schemes | dayofyear | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | {'001/GLDSMT_SERC_CalTarps_31July2017_am_l0s0/... | {'xmin': -76.5520244, 'ymin': 38.8724263, 'xma... | GLDSMT_001 | POLYGON ((-76.55112 38.87243, -76.54378 38.872... | GLDSMT_SERC_CalTarps_31July2017_am_l0s0 | [{'href': 'https://cmr.earthdata.nasa.gov/stac... | [https://stac-extensions.github.io/storage/v2.... | 1.1.0 | Feature | 2017-07-31 04:00:00+00:00 | 2017-08-01 03:59:59+00:00 | 2017-07-31 04:00:00+00:00 | {'aws': {'bucket': 'lp-prod-protected', 'platf... | 212 |
Now, let’s create a small areal subset of the data as we have for the other examples
c = gf_gliht.geometry.centroid
mini_aoi = gpd.GeoDataFrame(
geometry=[box(c.x - 0.0045, c.y - 0.0045, c.x + 0.0045, c.y + 0.0045)],
crs="EPSG:4326",
)
/tmp/ipykernel_1139/1428141286.py:1: UserWarning: Geometry is in a geographic CRS. Results from 'centroid' are likely incorrect. Use 'GeoSeries.to_crs()' to re-project geometries to a projected CRS before this operation.
c = gf_gliht.geometry.centroid
/home/docs/checkouts/readthedocs.org/user_builds/coincident/checkouts/104/.pixi/envs/dev/lib/python3.12/site-packages/shapely/geometry/polygon.py:91: FutureWarning: Calling float on a single element Series is deprecated and will raise a TypeError in the future. Use float(ser.iloc[0]) instead
return [float(c) for c in o]
m = gf_gliht.explore()
mini_aoi.explore(color="black", m=m)
As seen in the Additional Aerial LiDAR Datasets example, NASA G-LiHT has many different provided gridded datasets. The following collections below are the current datasets supported by coincident.
- 'ortho': Orthorectified aerial imagery
- 'chm': Canopy height model
- 'dsm': Digital surface model
- 'dtm': Digital terrain model
- 'hyperspectral_ancillary': Ancillary HSI data
- 'radiance': Hyperspectral aradiance
- 'reflectance': Hyperspectral surface reflectance
- 'hyperspectral_vegetation': HSI-derived veg indices
Note
Not all G-LiHT flights will contain every single product listed. For example, a flight may have ‘dsm’ data but not ‘ortho’ data.
# we'll need the dataset id from our initial search
gf_gliht.id.item()
'GLDSMT_SERC_CalTarps_31July2017_am_l0s0'
Important
Unlike the G-LiHT search, you will need NASA Earthdata credentials to read in and download the gridded datasets from G-LiHT. You can register for a free account here: https://urs.earthdata.nasa.gov/users/new
and set:
%%time
da_chm = coincident.io.xarray.load_gliht_raster(
aoi=mini_aoi,
dataset_id=gf_gliht.id.item(),
product="chm",
)
---------------------------------------------------------------------------
OSError Traceback (most recent call last)
File <timed exec>:1
File ~/checkouts/readthedocs.org/user_builds/coincident/checkouts/104/src/coincident/io/xarray.py:734, in load_gliht_raster(aoi, dataset_id, product)
728 if not username or not password:
729 msg_no_ed_cres = """
730 EARTHDATA_USER and EARTHDATA_PASS must be set in the environment.
731 os.environ["EARTHDATA_USER"] = "username"
732 os.environ["EARTHDATA_PASS"] ="password"
733 """
--> 734 raise OSError(msg_no_ed_cres)
736 # Use the href from the first found data asset to create the grid template
737 first_href = data_assets[asset_keys_to_load[0]]["href"]
OSError:
EARTHDATA_USER and EARTHDATA_PASS must be set in the environment.
os.environ["EARTHDATA_USER"] = "username"
os.environ["EARTHDATA_PASS"] ="password"
da_chm
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[65], line 1
----> 1 da_chm
NameError: name 'da_chm' is not defined
da_chm.plot.imshow(cmap="Greens");
Download#
Note
Since there are so many gridded products for G-LiHT, you can specify multiple products to download in one call. This download will automatically skip any products that aren’t available for the specific flight among the current datasets supported by coincident
coincident.io.download.download_gliht_raster(
aoi=mini_aoi,
dataset_id=gf_gliht.id.item(),
products=["chm", "dsm", "dtm"],
output_dir="/tmp",
)