Comparing Elevation Measurements#
Once you have an area of interest with multiple overlapping DEMs and/or laser altimetry measurements, you can quickly compare elevation values. coincident supports a main plotting panel for elevation comparisons, as well as dynamic boxplots and histograms for elevation differences over various terrain characteristics and land cover types.
This notebook highlights the following functions:
coincident.plot.compare_dems
coincident.plot.boxplot_slope
coincident.plot.boxplot_elevation
coincident.plot.boxplot_aspect
coincident.plot.hist_esa
Warning
Plotting routines are currently only tested for comparing datasets in EPSG:7912 (ITRF 2014)
import coincident
import geopandas as gpd
from shapely.geometry import box
import rioxarray as rxr
import numpy as np
import matplotlib.pyplot as plt
from rasterio.enums import Resampling
/home/docs/checkouts/readthedocs.org/user_builds/coincident/checkouts/104/src/coincident/io/download.py:27: TqdmExperimentalWarning: Using `tqdm.autonotebook.tqdm` in notebook mode. Use `tqdm.tqdm` instead to force console mode (e.g. in jupyter console)
from tqdm.autonotebook import tqdm
%matplotlib inline
# %config InlineBackend.figure_format = 'retina'
Load reference DEM#
workunit = "CO_WestCentral_2019"
df_wesm = coincident.search.wesm.read_wesm_csv()
gf_lidar = coincident.search.wesm.load_by_fid(
df_wesm[df_wesm.workunit == workunit].index
)
gf_lidar
| workunit | workunit_id | project | project_id | start_datetime | end_datetime | ql | spec | p_method | dem_gsd_meters | ... | seamless_category | seamless_reason | lpc_link | sourcedem_link | metadata_link | geometry | collection | datetime | dayofyear | duration | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | CO_WestCentral_2019 | 175984 | CO_WestCentral_2019_A19 | 175987 | 2019-08-21 | 2019-09-19 | QL 2 | USGS Lidar Base Specification 1.3 | linear-mode lidar | 1.0 | ... | Meets | Meets 3DEP seamless DEM requirements | https://rockyweb.usgs.gov/vdelivery/Datasets/S... | http://prd-tnm.s3.amazonaws.com/index.html?pre... | http://prd-tnm.s3.amazonaws.com/index.html?pre... | MULTIPOLYGON (((-106.17143 38.42061, -106.3208... | 3DEP | 2019-09-04 12:00:00 | 247 | 29 |
1 rows × 33 columns
# We will examine the 'sourcedem' 1m
# NOTE: it's important to note the *vertical* CRS of the data
gf_lidar.iloc[0]
workunit CO_WestCentral_2019
workunit_id 175984
project CO_WestCentral_2019_A19
project_id 175987
start_datetime 2019-08-21 00:00:00
end_datetime 2019-09-19 00:00:00
ql QL 2
spec USGS Lidar Base Specification 1.3
p_method linear-mode lidar
dem_gsd_meters 1.0
horiz_crs 6350
vert_crs 5703
geoid GEOID12B
lpc_pub_date 2021-01-30 00:00:00
lpc_update NaT
lpc_category Meets
lpc_reason Meets 3DEP LPC requirements
sourcedem_pub_date 2021-01-29 00:00:00
sourcedem_update NaT
sourcedem_category Meets
sourcedem_reason Meets 3DEP source DEM requirements
onemeter_category Meets
onemeter_reason Meets 3DEP 1-m DEM requirements
seamless_category Meets
seamless_reason Meets 3DEP seamless DEM requirements
lpc_link https://rockyweb.usgs.gov/vdelivery/Datasets/S...
sourcedem_link http://prd-tnm.s3.amazonaws.com/index.html?pre...
metadata_link http://prd-tnm.s3.amazonaws.com/index.html?pre...
geometry MULTIPOLYGON (((-106.171431 38.420609, -106.32...
collection 3DEP
datetime 2019-09-04 12:00:00
dayofyear 247
duration 29
Name: 0, dtype: object
# NOTE: since we're going to compare to satellite altimetry, let's convert first to EPSG:7912 !
# GDAL / PROJ will use the following transform by default
!projinfo -s EPSG:6350+5703 -t EPSG:7912 -q -o proj
+proj=pipeline
+step +inv +proj=aea +lat_0=23 +lon_0=-96 +lat_1=29.5 +lat_2=45.5 +x_0=0 +y_0=0
+ellps=GRS80
+step +proj=vgridshift +grids=us_noaa_g2018u0.tif +multiplier=1
+step +proj=cart +ellps=GRS80
+step +inv +proj=helmert +x=1.0053 +y=-1.90921 +z=-0.54157 +rx=0.02678138
+ry=-0.00042027 +rz=0.01093206 +s=0.00036891 +dx=0.00079 +dy=-0.0006
+dz=-0.00144 +drx=6.667e-05 +dry=-0.00075744 +drz=-5.133e-05
+ds=-7.201e-05 +t_epoch=2010 +convention=coordinate_frame
+step +inv +proj=cart +ellps=GRS80
+step +proj=unitconvert +xy_in=rad +xy_out=deg
+step +proj=axisswap +order=2,1
Now, let’s grab the vendor DEM products for two arbitrary tiles of this flight to look at elevation differences between the aerial LIDAR derved DEM, Copernicus GLO-30 DEM, ICESat-2 elevations, and GEDI elevations.
# We will rely on GDAL to do this reprojection
# gdalvsi = "/vsicurl?empty_dir=yes&url="
gdalvsi = "/vsicurl/"
url_tile_1 = f"{gdalvsi}https://prd-tnm.s3.amazonaws.com/StagedProducts/Elevation/OPR/Projects/CO_WestCentral_2019_A19/CO_WestCentral_2019/TIFF/USGS_OPR_CO_WestCentral_2019_A19_be_w1032n1780.tif"
url_tile_2 = f"{gdalvsi}https://prd-tnm.s3.amazonaws.com/StagedProducts/Elevation/OPR/Projects/CO_WestCentral_2019_A19/CO_WestCentral_2019/TIFF/USGS_OPR_CO_WestCentral_2019_A19_be_w1032n1779.tif"
# First build a mosaic in native 3D CRS
!gdalbuildvrt -q -a_srs EPSG:6350+5703 /tmp/mosaic.vrt '{url_tile_1}' '{url_tile_2}'
# Then reproject
!GDAL_DISABLE_READDIR_ON_OPEN=EMPTY_DIR gdalwarp -q -overwrite -t_srs EPSG:7912 -r bilinear /tmp/mosaic.vrt /tmp/mosaic_7912.tif
# Open our 'reference' DEM
da_lidar = rxr.open_rasterio("/tmp/mosaic_7912.tif", masked=True).squeeze()
da_lidar
<xarray.DataArray (y: 1917, x: 1465)> Size: 11MB
[2808405 values with dtype=float32]
Coordinates:
band int64 8B 1
* x (x) float64 12kB -108.0 -108.0 -108.0 ... -108.0 -108.0 -108.0
* y (y) float64 15kB 38.47 38.47 38.47 38.47 ... 38.46 38.46 38.46
spatial_ref int64 8B 0
Attributes:
AREA_OR_POINT: Area
scale_factor: 1.0
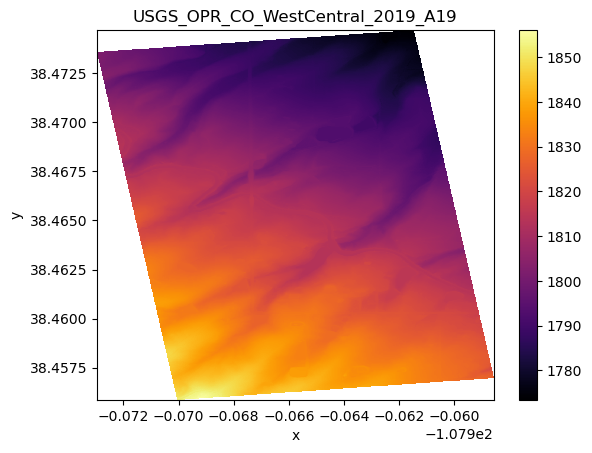
add_offset: 0.0# Plot a single DEM
ax = coincident.plot.plot_dem(da_lidar, title="USGS_OPR_CO_WestCentral_2019_A19");
Load secondary DEMs#
Note
Currently coincident facilitates loading select global DEMs in EPSG:7912, if your dataset is not in that CRS be sure to first reproject your data
# search geodataframe to grab the COP30 DEM
bbox_tile = da_lidar.rio.bounds()
poly_tile = box(*bbox_tile)
gf_tile = gpd.GeoDataFrame(geometry=[poly_tile], crs=da_lidar.rio.crs)
# NOTE: this is 30m data, but we are going to resample it to 1m to match our lidar
da_cop = coincident.io.xarray.load_dem_7912("cop30", aoi=gf_tile)
da_cop
---------------------------------------------------------------------------
KeyError Traceback (most recent call last)
File ~/checkouts/readthedocs.org/user_builds/coincident/checkouts/104/.pixi/envs/dev/lib/python3.12/site-packages/xarray/backends/file_manager.py:211, in CachingFileManager._acquire_with_cache_info(self, needs_lock)
210 try:
--> 211 file = self._cache[self._key]
212 except KeyError:
File ~/checkouts/readthedocs.org/user_builds/coincident/checkouts/104/.pixi/envs/dev/lib/python3.12/site-packages/xarray/backends/lru_cache.py:56, in LRUCache.__getitem__(self, key)
55 with self._lock:
---> 56 value = self._cache[key]
57 self._cache.move_to_end(key)
KeyError: [<function open at 0x710cb64256c0>, ('https://raw.githubusercontent.com/uw-cryo/3D_CRS_Transformation_Resources/refs/heads/main/globaldems/COP30_hh_7912.vrt',), 'r', (('sharing', False),), '3eea52ac-f7cd-4551-b944-1383aa69f64b']
During handling of the above exception, another exception occurred:
CPLE_OpenFailedError Traceback (most recent call last)
File rasterio/_base.pyx:310, in rasterio._base.DatasetBase.__init__()
File rasterio/_base.pyx:221, in rasterio._base.open_dataset()
File rasterio/_err.pyx:359, in rasterio._err.exc_wrap_pointer()
CPLE_OpenFailedError: '/vsicurl/https://raw.githubusercontent.com/uw-cryo/3D_CRS_Transformation_Resources/refs/heads/main/globaldems/COP30_hh_7912.vrt' does not exist in the file system, and is not recognized as a supported dataset name.
During handling of the above exception, another exception occurred:
RasterioIOError Traceback (most recent call last)
Cell In[11], line 2
1 # NOTE: this is 30m data, but we are going to resample it to 1m to match our lidar
----> 2 da_cop = coincident.io.xarray.load_dem_7912("cop30", aoi=gf_tile)
3 da_cop
File ~/checkouts/readthedocs.org/user_builds/coincident/checkouts/104/src/coincident/io/xarray.py:97, in load_dem_7912(dataset, aoi)
94 # Pull all data into memory
95 with ENV:
96 return (
---> 97 xr.open_dataarray(uri, engine="rasterio")
98 .rio.clip_box(**aoi.bounds)
99 .squeeze()
100 .load()
101 )
File ~/checkouts/readthedocs.org/user_builds/coincident/checkouts/104/.pixi/envs/dev/lib/python3.12/site-packages/xarray/backends/api.py:881, in open_dataarray(filename_or_obj, engine, chunks, cache, decode_cf, mask_and_scale, decode_times, decode_timedelta, use_cftime, concat_characters, decode_coords, drop_variables, inline_array, chunked_array_type, from_array_kwargs, backend_kwargs, **kwargs)
710 def open_dataarray(
711 filename_or_obj: str | os.PathLike[Any] | ReadBuffer | AbstractDataStore,
712 *,
(...) 731 **kwargs,
732 ) -> DataArray:
733 """Open an DataArray from a file or file-like object containing a single
734 data variable.
735
(...) 878 open_dataset
879 """
--> 881 dataset = open_dataset(
882 filename_or_obj,
883 decode_cf=decode_cf,
884 mask_and_scale=mask_and_scale,
885 decode_times=decode_times,
886 concat_characters=concat_characters,
887 decode_coords=decode_coords,
888 engine=engine,
889 chunks=chunks,
890 cache=cache,
891 drop_variables=drop_variables,
892 inline_array=inline_array,
893 chunked_array_type=chunked_array_type,
894 from_array_kwargs=from_array_kwargs,
895 backend_kwargs=backend_kwargs,
896 use_cftime=use_cftime,
897 decode_timedelta=decode_timedelta,
898 **kwargs,
899 )
901 if len(dataset.data_vars) != 1:
902 if len(dataset.data_vars) == 0:
File ~/checkouts/readthedocs.org/user_builds/coincident/checkouts/104/.pixi/envs/dev/lib/python3.12/site-packages/xarray/backends/api.py:687, in open_dataset(filename_or_obj, engine, chunks, cache, decode_cf, mask_and_scale, decode_times, decode_timedelta, use_cftime, concat_characters, decode_coords, drop_variables, inline_array, chunked_array_type, from_array_kwargs, backend_kwargs, **kwargs)
675 decoders = _resolve_decoders_kwargs(
676 decode_cf,
677 open_backend_dataset_parameters=backend.open_dataset_parameters,
(...) 683 decode_coords=decode_coords,
684 )
686 overwrite_encoded_chunks = kwargs.pop("overwrite_encoded_chunks", None)
--> 687 backend_ds = backend.open_dataset(
688 filename_or_obj,
689 drop_variables=drop_variables,
690 **decoders,
691 **kwargs,
692 )
693 ds = _dataset_from_backend_dataset(
694 backend_ds,
695 filename_or_obj,
(...) 705 **kwargs,
706 )
707 return ds
File ~/checkouts/readthedocs.org/user_builds/coincident/checkouts/104/.pixi/envs/dev/lib/python3.12/site-packages/rioxarray/xarray_plugin.py:59, in RasterioBackend.open_dataset(self, filename_or_obj, drop_variables, parse_coordinates, lock, masked, mask_and_scale, variable, group, default_name, decode_coords, decode_times, decode_timedelta, band_as_variable, open_kwargs)
57 if open_kwargs is None:
58 open_kwargs = {}
---> 59 rds = _io.open_rasterio(
60 filename_or_obj,
61 parse_coordinates=parse_coordinates,
62 cache=False,
63 lock=lock,
64 masked=masked,
65 mask_and_scale=mask_and_scale,
66 variable=variable,
67 group=group,
68 default_name=default_name,
69 decode_times=decode_times,
70 decode_timedelta=decode_timedelta,
71 band_as_variable=band_as_variable,
72 **open_kwargs,
73 )
74 if isinstance(rds, xarray.DataArray):
75 dataset = rds.to_dataset()
File ~/checkouts/readthedocs.org/user_builds/coincident/checkouts/104/.pixi/envs/dev/lib/python3.12/site-packages/rioxarray/_io.py:1135, in open_rasterio(filename, parse_coordinates, chunks, cache, lock, masked, mask_and_scale, variable, group, default_name, decode_times, decode_timedelta, band_as_variable, **open_kwargs)
1133 else:
1134 manager = URIManager(file_opener, filename, mode="r", kwargs=open_kwargs)
-> 1135 riods = manager.acquire()
1136 captured_warnings = rio_warnings.copy()
1138 # raise the NotGeoreferencedWarning if applicable
File ~/checkouts/readthedocs.org/user_builds/coincident/checkouts/104/.pixi/envs/dev/lib/python3.12/site-packages/xarray/backends/file_manager.py:193, in CachingFileManager.acquire(self, needs_lock)
178 def acquire(self, needs_lock=True):
179 """Acquire a file object from the manager.
180
181 A new file is only opened if it has expired from the
(...) 191 An open file object, as returned by ``opener(*args, **kwargs)``.
192 """
--> 193 file, _ = self._acquire_with_cache_info(needs_lock)
194 return file
File ~/checkouts/readthedocs.org/user_builds/coincident/checkouts/104/.pixi/envs/dev/lib/python3.12/site-packages/xarray/backends/file_manager.py:217, in CachingFileManager._acquire_with_cache_info(self, needs_lock)
215 kwargs = kwargs.copy()
216 kwargs["mode"] = self._mode
--> 217 file = self._opener(*self._args, **kwargs)
218 if self._mode == "w":
219 # ensure file doesn't get overridden when opened again
220 self._mode = "a"
File ~/checkouts/readthedocs.org/user_builds/coincident/checkouts/104/.pixi/envs/dev/lib/python3.12/site-packages/rasterio/env.py:463, in ensure_env_with_credentials.<locals>.wrapper(*args, **kwds)
460 session = DummySession()
462 with env_ctor(session=session):
--> 463 return f(*args, **kwds)
File ~/checkouts/readthedocs.org/user_builds/coincident/checkouts/104/.pixi/envs/dev/lib/python3.12/site-packages/rasterio/__init__.py:356, in open(fp, mode, driver, width, height, count, crs, transform, dtype, nodata, sharing, opener, **kwargs)
353 path = _parse_path(raw_dataset_path)
355 if mode == "r":
--> 356 dataset = DatasetReader(path, driver=driver, sharing=sharing, **kwargs)
357 elif mode == "r+":
358 dataset = get_writer_for_path(path, driver=driver)(
359 path, mode, driver=driver, sharing=sharing, **kwargs
360 )
File rasterio/_base.pyx:312, in rasterio._base.DatasetBase.__init__()
RasterioIOError: '/vsicurl/https://raw.githubusercontent.com/uw-cryo/3D_CRS_Transformation_Resources/refs/heads/main/globaldems/COP30_hh_7912.vrt' does not exist in the file system, and is not recognized as a supported dataset name.
ds_cop_r = (
da_cop.rio.reproject_match(
da_lidar,
resampling=Resampling.bilinear,
)
.where(da_lidar.notnull())
.to_dataset(name="elevation")
)
ds_cop_r
# Create hillshade variables for plot backgrounds
# This function expects Datasets
ds_lidar = da_lidar.to_dataset(name="elevation")
ds_lidar["hillshade"] = coincident.plot.gdaldem(
ds_lidar.elevation, "hillshade", scale=111120
)
ds_cop_r["hillshade"] = coincident.plot.gdaldem(
ds_cop_r.elevation, "hillshade", scale=111120
)
# Plot a single DEM with hillshade
ax = coincident.plot.plot_dem(
ds_cop_r["elevation"],
title="Resampled Copernicus DEM",
da_hillshade=ds_lidar["hillshade"],
)
# Get GEDI
gf_gedi = coincident.search.search(
dataset="gedi",
intersects=gf_tile,
)
data_gedi = coincident.io.sliderule.subset_gedi02a(
gf_gedi, gf_tile, include_worldcover=True
)
# Get ICESAT-2
gf_is2 = coincident.search.search(
dataset="icesat-2",
intersects=gf_tile,
)
data_is2 = coincident.io.sliderule.subset_atl06(
gf_is2, gf_tile, include_worldcover=True
)
# NOTE: this works because ICESat-2 and GEDI are EPSG:7912
coincident.plot.plot_altimeter_points(
data_is2, "h_li", da_hillshade=ds_lidar["hillshade"], title="ICESat-2 ATL06 points"
);
Main Elevation Comparison Panel#
‘coincident’ supports a main compare_dems function that allows you to compare multiple DEMs and altimetry points in a single panel. This function dynamically adjusts based on the number of datasets provided.
Note
compare_dems assumes that all DEMs are in the same coordinate reference system (CRS) and aligned. It also assumes that each DEM has an ‘elevation’ variable. If add_hillshade=True, it assumes the first DEM passed has a ‘hillshade’ variable, where the ‘hillshade’ variable can be calculated with coincident.plot.create_hillshade as seen below. A minimum of two DEMs must be provided and a maximum of 5 total datasets (datasets being DEMs and GeoDataFrames of altimetry points) can be provided.
# list of DEMs to compare, where the first DEM in the list is the 'source' DEM
dem_list = [ds_lidar, ds_cop_r]
# dictionary of GeoDataFrames with altimetry points to compare
# where the key is the name/plot title of the dataset and the value is a tuple of the respective
# GeoDataFrame and the column name of the elevation values of interest
dicts_gds = {"ICESat-2": (data_is2, "h_li"), "GEDI": (data_gedi, "elevation_lm")}
axd = coincident.plot.compare_dems(
dem_list,
dicts_gds,
# ds_wc=ds_wc,
add_hillshade=True,
diff_clim=(-5, 5), # difference colormap limits
)
Dynamic Elevation Difference Boxplots Based on Terrain Variables (slope, elevation, aspect)#
coincident allows users to dynamically plot elevation differences based on grouped data such as slope values. In these functions (that are wrappers for the generalized coincident.plot.boxplot_terrain_diff function), the user provides a reference DEM and one other DEM or GeoDataFrame with elevation values to compare to. These function assumes that the reference DEM has the respective terrain variable (slope, aspect, etc.) of interest.
Note
Boxplots will only be generated with groups of value counts greater than 30. Dynamic ylims will be fit to the IQRs of the boxplots.
# These routines assume rectilinear data, so we first convert to UTM
crs_utm = ds_lidar.rio.estimate_utm_crs()
ds_lidar_utm = ds_lidar.elevation.rio.reproject(crs_utm).to_dataset()
ds_cop_utm = ds_cop_r.elevation.rio.reproject(crs_utm).to_dataset()
slope = coincident.plot.utils.gdaldem(ds_lidar_utm.elevation, "slope")
ds_lidar_utm["slope"] = slope
_, ax = plt.subplots()
ds_lidar_utm.slope.plot.imshow(ax=ax)
ax.set_title("USGS_OPR_CO_WestCentral_2019_A19");
ax_boxplot_dems = coincident.plot.boxplot_slope([ds_lidar_utm, ds_cop_utm])
# we also support elevation differences based on elevation bins
# grouped on the reference DEM's elevation values
ax_elev = coincident.plot.boxplot_elevation(
[ds_lidar_utm, ds_cop_utm], elevation_bins=np.arange(1700, 2000, 20)
)
# lastly for the boxplots, let's look at aspect
aspect = coincident.plot.utils.gdaldem(ds_lidar_utm.elevation, "aspect")
ds_lidar_utm["aspect"] = aspect
_, ax = plt.subplots()
ds_lidar_utm.aspect.plot.imshow(ax=ax)
ax.set_title("USGS_OPR_CO_WestCentral_2019_A19");
ax_aspect = coincident.plot.boxplot_aspect([ds_lidar_utm, ds_cop_utm])
Histograms of Elevation Differences over Land Cover#
Plot elevation differences between DEMs or point data, grouped by ESA World Cover 2020 land cover class
Note
Histograms will only be generated with groups of value counts greater than a user-defined threshold (default is 30).
ax_lulc_cop30 = coincident.plot.hist_esa([ds_lidar, ds_cop_r])