Sliderule data I/O#
This notebook will highlight analyzing various coincident elevation measurements. We will find regions with and use slideruleearth.io service to retrieve ICESat-2 and GEDI point elevation measurements.
Note
Keep in mind, these measurements are from different sensor types, close in time, but not at exactly the same time. All measurements also have uncertainties, so we do not expect perfect agreement.
import coincident
from mpl_toolkits.axes_grid1.inset_locator import inset_axes
import matplotlib.pyplot as plt
import numpy as np
# For testing
# import sliderule
# sliderule.init(url='slideruleearth.io', verbose=True)
/home/docs/checkouts/readthedocs.org/user_builds/coincident/checkouts/104/src/coincident/io/download.py:27: TqdmExperimentalWarning: Using `tqdm.autonotebook.tqdm` in notebook mode. Use `tqdm.tqdm` instead to force console mode (e.g. in jupyter console)
from tqdm.autonotebook import tqdm
%matplotlib inline
# %config InlineBackend.figure_format = 'retina'
Identify a primary dataset#
Start by loading a full resolution polygon of a 3DEP LiDAR workunit which has a known start_datetime and end_datatime:
workunit = "CO_WestCentral_2019"
df_wesm = coincident.search.wesm.read_wesm_csv()
gf_lidar = coincident.search.wesm.load_by_fid(
df_wesm[df_wesm.workunit == workunit].index
)
gf_lidar
| workunit | workunit_id | project | project_id | start_datetime | end_datetime | ql | spec | p_method | dem_gsd_meters | ... | seamless_category | seamless_reason | lpc_link | sourcedem_link | metadata_link | geometry | collection | datetime | dayofyear | duration | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | CO_WestCentral_2019 | 175984 | CO_WestCentral_2019_A19 | 175987 | 2019-08-21 | 2019-09-19 | QL 2 | USGS Lidar Base Specification 1.3 | linear-mode lidar | 1.0 | ... | Meets | Meets 3DEP seamless DEM requirements | https://rockyweb.usgs.gov/vdelivery/Datasets/S... | http://prd-tnm.s3.amazonaws.com/index.html?pre... | http://prd-tnm.s3.amazonaws.com/index.html?pre... | MULTIPOLYGON (((-106.17143 38.42061, -106.3208... | 3DEP | 2019-09-04 12:00:00 | 247 | 29 |
1 rows × 33 columns
Search secondary datasets#
Provide a list that will be searched in order. The list contains tuples of dataset aliases and the temporal pad in days to search before the primary dataset start and end dates
secondary_datasets = [
("gedi", 40), # +/- 40 days from lidar
("icesat-2", 60), # +/- 60 days from lidar
]
gf_gedi, gf_is2 = coincident.search.cascading_search(
gf_lidar,
secondary_datasets,
min_overlap_area=30, # km^2
)
Get ICESat-2 ATL06 Data#
We’ve identified 7 granules of icesat-2 data to examine, but there is no need to work with the entire granule, which spans a huge geographic extent. Instead we’ll retrieve a subset of elevation values in the area of interest for each granule.
# Hone in on single granule as a pandas series
i = 0
granule_gdf = gf_is2.iloc[[i]]
granule = gf_is2.iloc[i]
granule_gdf.plot() # needs to be a geodataframe
plt.title(granule.id);
# The icesat-2 track trends N-S, and is crossed by multiple GEDI tracks, resulting in the crosshatched appearance
data_is2 = coincident.io.sliderule.subset_atl06(
granule_gdf,
include_worldcover=True,
)
# Plot this data and overlay footprint
fig, ax = plt.subplots(figsize=(8, 10))
plt.scatter(
x=data_is2.geometry.x,
y=data_is2.geometry.y,
c=data_is2.h_li,
s=10,
)
granule_gdf.dissolve().boundary.plot(ax=ax, color="magenta")
cb = plt.colorbar()
cb.set_label("elevation_hr (m)")
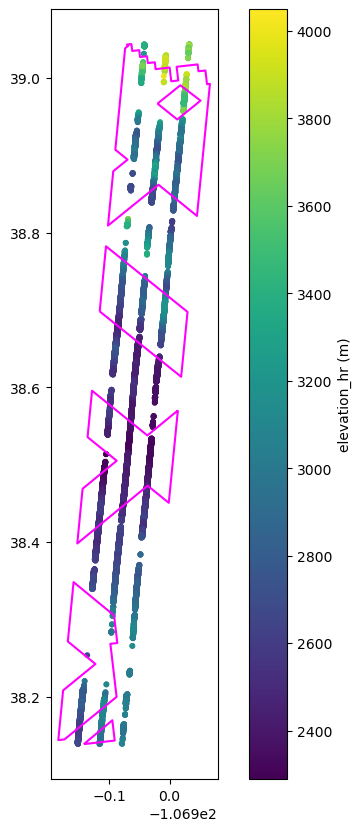
Note
sliderule gets data from the envelope of multipolygon, not only data within intersecting GEDI tracks. Along-track gaps are where there is missing data.
# Here, we specify aoi=granule_gdf to only return GEDI data
# in our icesat-2 track of interest
data_gedi = coincident.io.sliderule.subset_gedi02a(
gf_gedi,
aoi=granule_gdf,
include_worldcover=True,
)
data_gedi
| solar_elevation | flags | sensitivity | elevation_hr | elevation_lm | beam | track | orbit | geometry | worldcover.time | worldcover.value | worldcover.flags | worldcover.file_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| time | |||||||||||||
| 2019-09-23 08:03:40.028272128 | -48.950699 | 130 | 0.915599 | 2970.799561 | 2964.996582 | 2 | 480 | 4416 | POINT (-107.05912 38.62987) | 1.309046e+09 | Tree cover | 0.0 | 4.724464e+10 |
| 2019-09-23 08:03:40.065466880 | -48.957371 | 130 | 0.935781 | 3084.797852 | 3060.687012 | 3 | 480 | 4416 | POINT (-107.05798 38.62212) | 1.309046e+09 | Tree cover | 0.0 | 4.724464e+10 |
| 2019-09-23 08:03:40.148106752 | -48.958637 | 130 | 0.900905 | 3043.598877 | 3021.360107 | 3 | 480 | 4416 | POINT (-107.0529 38.61885) | 1.309046e+09 | Tree cover | 0.0 | 4.724464e+10 |
| 2019-09-23 08:03:40.164634624 | -48.958889 | 130 | 0.906498 | 3023.866211 | 3002.188965 | 3 | 480 | 4416 | POINT (-107.05188 38.6182) | 1.309046e+09 | Tree cover | 0.0 | 4.724464e+10 |
| 2019-09-23 08:03:40.164634624 | -48.960491 | 130 | 0.905244 | 2792.681885 | 2769.727051 | 5 | 480 | 4416 | POINT (-107.07904 38.62588) | 1.309046e+09 | Tree cover | 0.0 | 4.724464e+10 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 2019-10-27 11:45:12.254580224 | -21.202047 | 130 | 0.960664 | 3284.958740 | 3254.115967 | 11 | 2006 | 4946 | POINT (-107.06034 39.0425) | 1.309046e+09 | Tree cover | 0.0 | 8.160438e+10 |
| 2019-10-27 11:45:12.262844416 | -21.201624 | 130 | 0.962776 | 3294.617432 | 3278.242676 | 11 | 2006 | 4946 | POINT (-107.05984 39.04281) | 1.309046e+09 | Grassland | 0.0 | 8.160438e+10 |
| 2019-10-27 11:45:12.271110400 | -21.201200 | 130 | 0.967792 | 3308.820557 | 3302.016357 | 11 | 2006 | 4946 | POINT (-107.05934 39.04312) | 1.309046e+09 | Grassland | 0.0 | 8.160438e+10 |
| 2019-10-27 11:45:12.279376384 | -21.200781 | 130 | 0.909410 | 3327.383057 | 3322.971680 | 11 | 2006 | 4946 | POINT (-107.05884 39.04343) | 1.309046e+09 | Grassland | 0.0 | 8.160438e+10 |
| 2019-10-27 11:45:12.287642368 | -21.200354 | 130 | 0.972252 | 3328.869141 | 3320.681641 | 11 | 2006 | 4946 | POINT (-107.05832 39.04376) | NaN | NaN | NaN | NaN |
7195 rows × 13 columns
# Plot this data and overlay footprint
fig, ax = plt.subplots(figsize=(8, 10))
plt.scatter(
x=data_gedi.geometry.x,
y=data_gedi.geometry.y,
c=data_gedi.elevation_hr,
s=10,
)
granule_gdf.dissolve().boundary.plot(ax=ax, color="magenta")
cb = plt.colorbar()
cb.set_label("elevation_hr (m)")
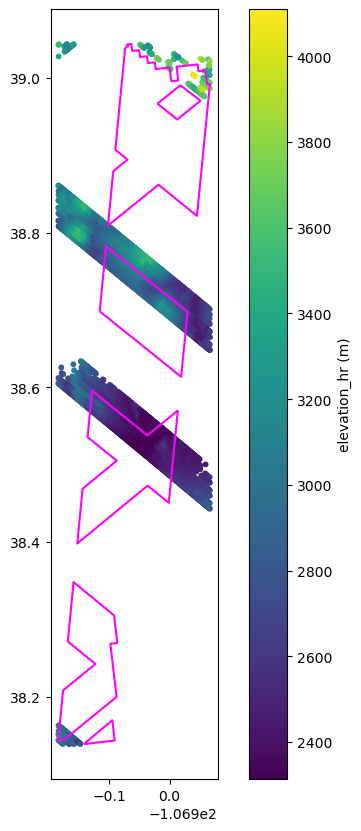
NOTE Like ICESat-2, it’s possible to have points falling outside the estimated footprints
Spatial join: nearest points#
NOTE: we will not worry about the difference in time of acquisition between adjacent points for now
utm_crs = granule_gdf.estimate_utm_crs()
data_is2_utm = data_is2.to_crs(utm_crs)
data_gedi_utm = data_gedi.to_crs(utm_crs)
# find nearest IS2 point to each GEDI
close_gedi = data_gedi_utm.sjoin_nearest(
data_is2_utm,
how="left",
max_distance=100, # at most 100m apart
distance_col="distances",
)
# index is GEDI subset (with _left added), with _right appended to is2 columns + Distances in meters
close_gedi = close_gedi[close_gedi["distances"].notna()]
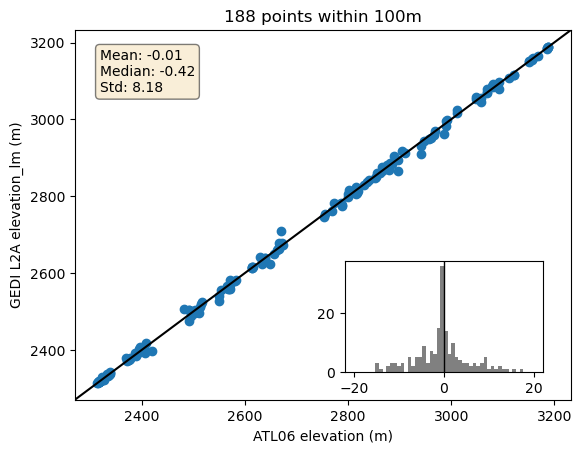
fig, ax = plt.subplots()
plt.scatter(close_gedi.h_li, close_gedi.elevation_lm)
ax.axline((0, 0), slope=1, color="k", transform=ax.transAxes)
plt.xlabel("ATL06 elevation (m)")
plt.ylabel("GEDI L2A elevation_lm (m)")
plt.title(f"{len(close_gedi)} points within 100m")
# Add statistics and inset histogram to plot
inset_ax = inset_axes(ax, width="40%", height="30%", borderpad=2, loc="lower right")
residuals = close_gedi.h_li - close_gedi.elevation_lm
mean_residual = np.mean(residuals)
median_residual = np.median(residuals)
std_residual = np.std(residuals)
textstr = "\n".join(
(
f"Mean: {mean_residual:.2f}",
f"Median: {median_residual:.2f}",
f"Std: {std_residual:.2f}",
)
)
props = dict(boxstyle="round", facecolor="wheat", alpha=0.5)
ax.text(
0.05,
0.95,
textstr,
transform=ax.transAxes,
fontsize=10,
verticalalignment="top",
bbox=props,
)
_ = inset_ax.hist(residuals, bins=50, range=(-20, 20), color="gray")
inset_ax.axvline(0, color="k", linestyle="-", linewidth=1);
Sample 3DEP#
Sample 3DEP 1m DEM at the subset of GEDI Points
Note
SlideRule returns all elevation data in EPSG:7912
close_gedi_3dep = coincident.io.sliderule.sample_3dep(
close_gedi,
# Restrict to only DEMs derived from specific WESM LiDAR project
project_name=gf_lidar["project"].iloc[0],
)
# Add 3DEP elevation values to existing dataframe
close_gedi["elevation_3dep"] = close_gedi_3dep.value.values
# Geometries of nearest IS2 points
close_is2 = data_is2_utm.loc[close_gedi.time_right]
# Sample 3DEP at the subset of GEDI Points
close_is2_3dep = coincident.io.sliderule.sample_3dep(
close_is2,
project_name=gf_lidar["project"].iloc[0],
)
close_is2["elevation_3dep"] = close_is2_3dep.value.values
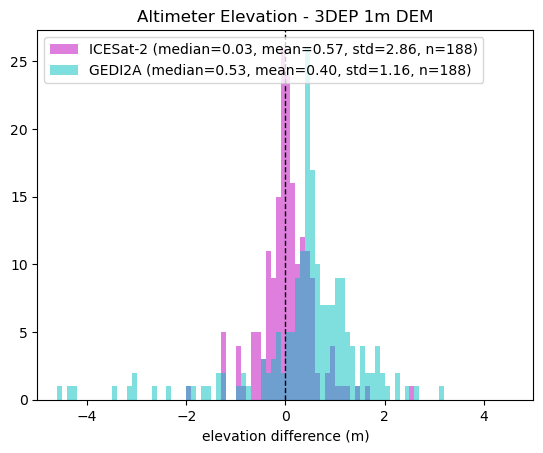
plt.axvline(0, color="k", linestyle="dashed", linewidth=1)
diff_3dep = close_is2.h_li.values - close_is2.elevation_3dep
label = f"ICESat-2 (median={np.nanmedian(diff_3dep):.2f}, mean={np.nanmean(diff_3dep):.2f}, std={np.nanstd(diff_3dep):.2f}, n={len(diff_3dep)})"
plt.hist(diff_3dep, range=[-5, 5], bins=100, color="m", alpha=0.5, label=label)
diff_3dep = close_gedi.elevation_lm.values - close_gedi.elevation_3dep
label = f"GEDI2A (median={np.nanmedian(diff_3dep):.2f}, mean={np.nanmean(diff_3dep):.2f}, std={np.nanstd(diff_3dep):.2f}, n={len(diff_3dep)})"
plt.hist(diff_3dep, range=[-5, 5], bins=100, color="c", alpha=0.5, label=label)
plt.xlabel("elevation difference (m)")
plt.title("Altimeter Elevation - 3DEP 1m DEM")
plt.xlim(-5, 5)
plt.legend(loc="upper left");